男女共同参画学協会連絡会
支援企業による広告記事
- ソーラボジャパン株式会社
- CAD を使って光学装置を設計してみよう
- 「生物物理」2025年10月号
- ソーラボジャパン株式会社
- 次世代2光子顕微鏡―小型化がもたらす新たな可能性
- 「生物物理」2024年10月号
- ソーラボジャパン株式会社
- サイエンティフィックカメラと周辺機器の同期
- 「生物物理」2023年10月号
- ソーラボジャパン株式会社
- 顕微鏡のリノベーション ~ 顕微鏡ポートを活用した光学系の導入
- 「生物物理」2022年12月号
「Biophysics and Physicobiology」に Junichi Higo, Kota Kasahara, Shun Sakuraba, Gert-Jan Bekker, Narutoshi Kamiya, Ikuo Fukuda, Takuya Takahashi, Yoshifumi Fukunishi による "A virtual system-coupled molecular dynamics simulation free from experimental knowledge on binding sites: Application to RNA-ligand binding free-energy landscape" をJ-STAGEの早期公開版として掲載
2025年04月29日 学会誌
日本生物物理学会欧文誌[Biophysics and Physicobiology]に以下の論文が早期公開されました。
Junichi Higo, Kota Kasahara, Shun Sakuraba, Gert-Jan Bekker, Narutoshi Kamiya, Ikuo Fukuda, Takuya Takahashi, Yoshifumi Fukunishi
"A virtual system-coupled molecular dynamics simulation free from experimental knowledge on binding sites: Application to RNA-ligand binding free-energy landscape"
URL:https://doi.org/10.2142/biophysico.bppb-v22.0011
- Abstract
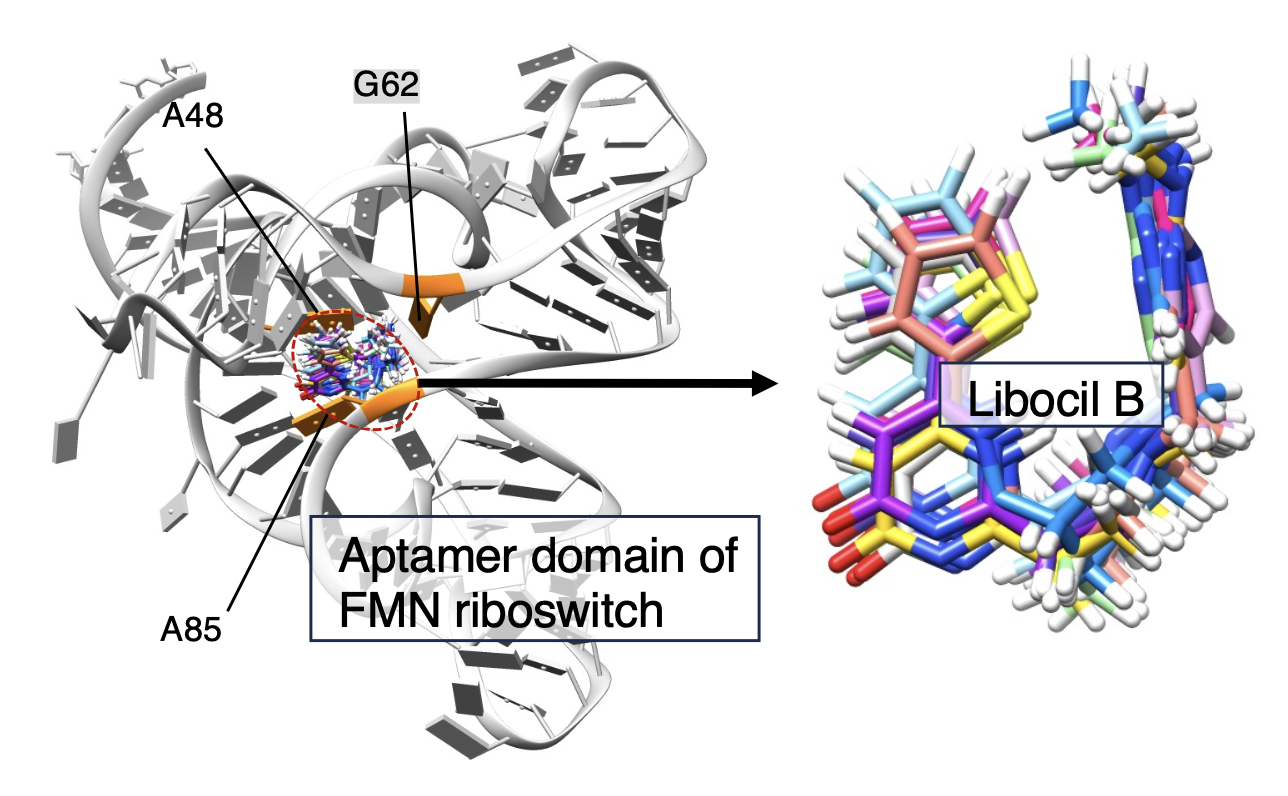
- Ligand–receptor docking simulation is difficult when the biomolecules have high intrinsic flexibility. If some knowledge on the ligand–receptor complex structure or inter-molecular contact sites are presented in advance, the difficulty of docking problem considerably decreases. This paper proposes a generalized-ensemble method “cartesian-space division mD-VcMD” (or CSD-mD-VcMD), which calculates stable complex structures without assist of experimental knowledge on the complex structure. This method is an extension of our previous method that requires the knowledge on the ligand–receptor complex structure in advance. Both the present and previous methods enhance the conformational sampling, and finally produce a binding free-energy landscape starting from a completely dissociated conformation, and provide a free-energy landscape. We applied the present method to same system studied by the previous method: A ligand (ribocil A or ribocil B) binding to an RNA (the aptamer domain of the FMN riboswitch). The two methods produced similar results, which explained experimental data. For instance, ribocil B bound to the aptamer’s deep binding pocket more strongly than ribocil A did. However, this does not mean that two methods have a similar performance. Note that the present method did not use the experimental knowledge of binding sites although the previous method was supported by the knowledge. The RNA-ligand binding site could be a cryptic site because RNA and ligand are highly flexible in general. The current study showed that CSD-mD-VcMD is actually useful to obtain a binding free-energy landscape of a flexible system, i.e., the RNA-ligand interacting system.
URL: https://doi.org/10.2142/biophysico.bppb-v22.0011