男女共同参画学協会連絡会
支援企業による広告記事
- ソーラボジャパン株式会社
- CAD を使って光学装置を設計してみよう
- 「生物物理」2025年10月号
- ソーラボジャパン株式会社
- 次世代2光子顕微鏡―小型化がもたらす新たな可能性
- 「生物物理」2024年10月号
- ソーラボジャパン株式会社
- サイエンティフィックカメラと周辺機器の同期
- 「生物物理」2023年10月号
- ソーラボジャパン株式会社
- 顕微鏡のリノベーション ~ 顕微鏡ポートを活用した光学系の導入
- 「生物物理」2022年12月号
「Biophysics and Physicobiology」に Daisuke Kohda, Seiichiro Hayashi, Daisuke Fujinami による "Residue-based correlation between equilibrium and rate constants is an experimental formulation of the consistency principle for smooth structural changes of proteins" をJ-STAGEの早期公開版として掲載
2023年12月13日 学会誌
日本生物物理学会欧文誌[Biophysics and Physicobiology]に以下の論文が早期公開されました。
Daisuke Kohda, Seiichiro Hayashi, Daisuke Fujinami
"Residue-based correlation between equilibrium and rate constants is an experimental formulation of the consistency principle for smooth structural changes of proteins"
URL:https://doi.org/10.2142/biophysico.bppb-v20.0046
- Abstract
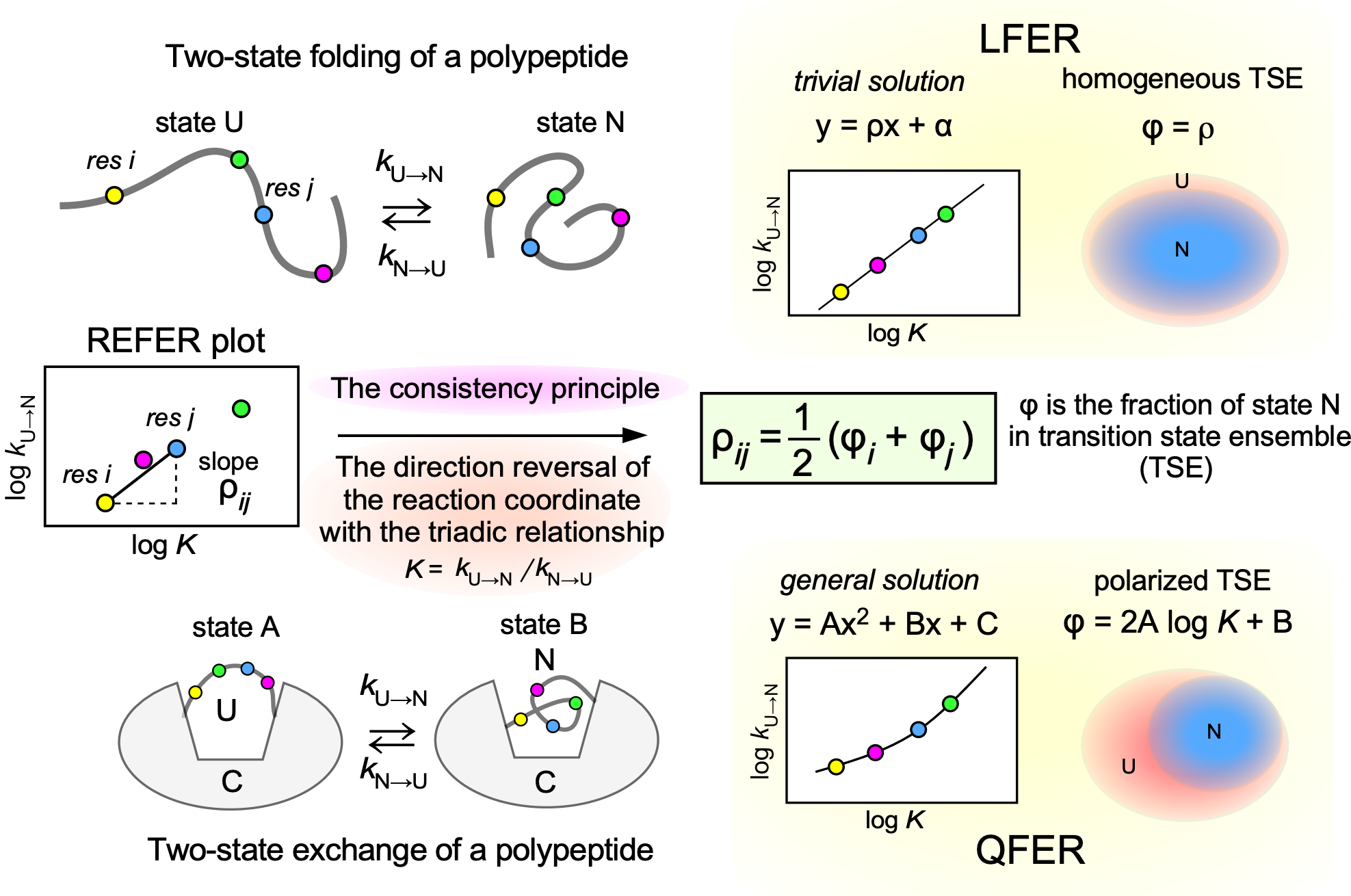
- The consistency principle represents a physicochemical condition requisite for ideal protein folding. It assumes that any pair of amino acid residues in partially folded structures has an attractive short-range interaction only if the two residues are in contact within the native structure. The residue-specific equilibrium constant, K, and the residue-specific rate constant, k (forward and backward), can be determined by NMR and hydrogen-deuterium exchange studies. Linear free energy relationships (LFER) in the rate-equilibrium free energy relationship (REFER) plots (i.e., log k vs. log K) are widely seen in protein-related phenomena, but our REFER plot differs from them in that the data points are derived from one polypeptide chain under a single condition. Here, we examined the theoretical basis of the residue-based LFER. First, we derived a basic equation, ρij = ½(φi + φj), from the consistency principle, where ρij is the slope of the line segment that connects residues i and j in the REFER plot, and φi and φj are the local fractions of the native state in the transient state ensemble (TSE). Next, we showed that the general solution is the alignment of the (log K, log k) data points on a parabolic curve in the REFER plot. Importantly, unlike LFER, the quadratic free energy relationship (QFER) is compatible with the heterogeneous formation of local structures in the TSE. Residue-based LFER/QFER provides a unique insight into the TSE: A foldable polypeptide chain consists of several folding units, which are consistently coupled to undergo smooth structural changes.
URL: https://doi.org/10.2142/biophysico.bppb-v20.0046