1
"MOLASS: Software for automatic processing of matrix data obtained from small-angle X-ray scattering and UV–visible spectroscopy combined with size-exclusion chromatography" by Kento Yonezawa, Masatsuyo Takahashi, Keiko Yatabe, Yasuko Nagatani, Nobutaka Shimizu is published in BPPB as the J-STAGE Advance Publication.
2023 January 07 BPPB
A following article is published as the J-STAGE Advance Publication in "Biophysics and Physicobiology".
Kento Yonezawa, Masatsuyo Takahashi, Keiko Yatabe, Yasuko Nagatani, Nobutaka Shimizu
"MOLASS: Software for automatic processing of matrix data obtained from small-angle X-ray scattering and UV–visible spectroscopy combined with size-exclusion chromatography"
URL:https://doi.org/10.2142/biophysico.bppb-v20.0001
- Abstract
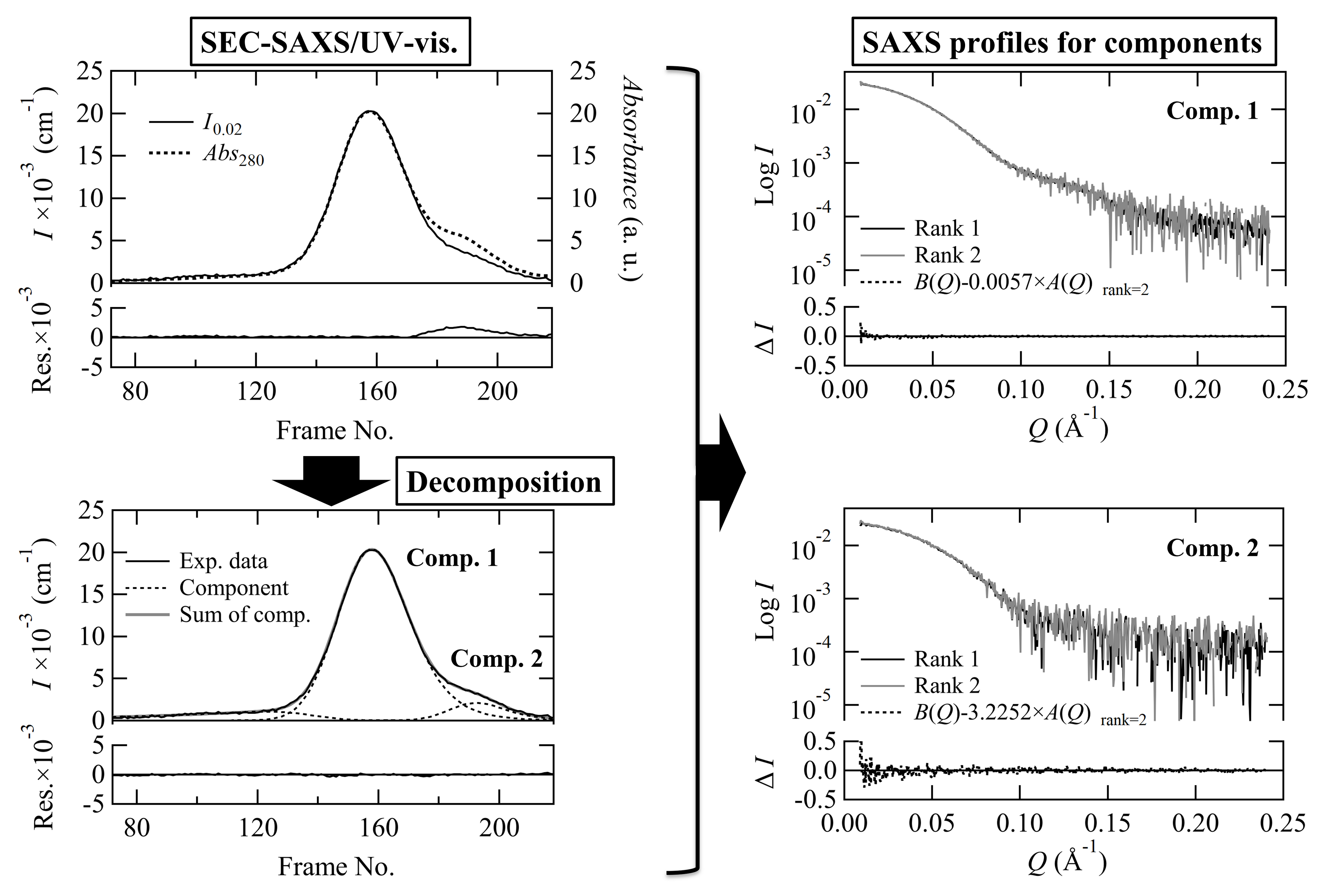
- Recent small-angle X-ray scattering (SAXS) for biological macromolecules (BioSAXS) is generally combined with size-exclusion chromatography (SEC-SAXS) at synchrotron facilities worldwide. For SEC-SAXS analysis, the final scattering profile for the target molecule is calculated from a large volume of continuously collected data. It would be ideal to automate this process; however, several complex problems exist regarding data measurement and analysis that have prevented automation. Here, we developed the analytical software MOLASS (Matrix Optimization with Low-rank factorization for Automated analysis of SEC-SAXS) to automatically calculate the final scattering profiles for solution structure analysis of target molecules. In this paper, the strategies for automatic analysis of SEC-SAXS data are described, including correction of baseline-drift using a low percentile method, optimization of peak decompositions composed of multiple scattering components using modified Gaussian fitting against the chromatogram, and rank determination for extrapolation to infinite dilution. In order to easily calculate each scattering component, the Moore-Penrose pseudo-inverse matrix is adopted as a basic calculation. Furthermore, this analysis method, in combination with UV–visible spectroscopy, led to better results in terms of accuracy in peak decomposition. Therefore, MOLASS will be able to smoothly suggest to users an accurate scattering profile for the subsequent structural analysis.